Before Ferhat Ay, Ph.D., entered first grade, he had already announced to the world that he wanted to become a computer engineer. By the time he enrolled at the Middle East Technical University in Ankara, Turkey, the avid gamer had narrowed his career path to video game developer and soon was busy taking math and computer graphics classes. In the midst of his undergraduate studies, an inspiring bioinformatics class captured his imagination and Dr. Ay found himself veering off into a new direction. Instead of dreaming up fantasy worlds, he redirected his focus to the string of four letters that make up our DNA.

“You are playing with A, T, G, and Cs and they all have meaning. You can look at them in very creative ways,” says Dr. Ay, who joined La Jolla Institute as Institute Leadership Assistant Professor of Computational Biology a little more than a year ago. “When I applied to Ph.D. programs, I specifically sought out bioinformatics programs and then it picked up from there.”
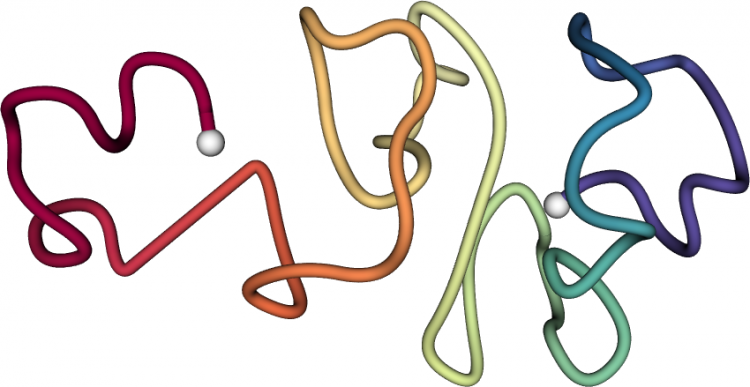
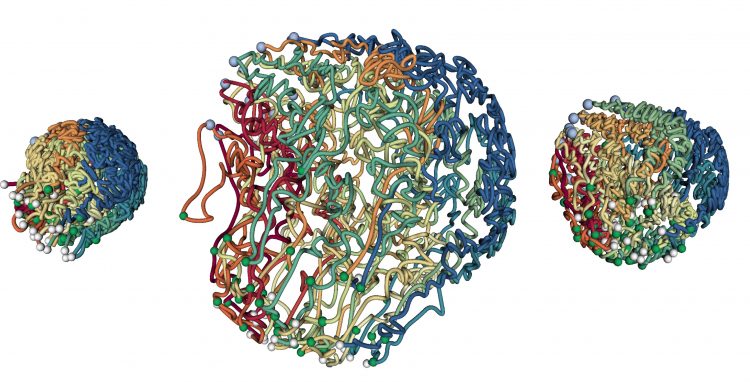
When fully extended, the DNA encoding our genes measures about two yards, yet it is tucked into a cell nucleus with a diameter more than a 100,000 times smaller. To fit it all in, the DNA is coiled up and carefully folded. Dr. Ay’s trailblazing work in the parasite Plasmodium falciparum, which causes the most lethal form of malaria, provided the first evidence that the three-dimensional shape of the resulting DNA folds can coordinately repress or activate genes.
“It was great to be in the right lab at the right time and be part of it when it all started,” he says. Since then the field of genome architecture has taken off and the genome’s three-dimensional structure has emerged as a universal tool to understand the regulation of gene function. At La Jolla Institute, Dr. Ay has expanded his research to uncover new links between disrupted genome folding patterns and disease.
HOW WOULD YOU DESCRIBE THE FOCUS OF YOUR RESEARCH?
I am interested in epigenetics. By definition, epigenetics is anything beyond the sequence of the four letters in DNA that controls the activity of genes. That would be chemical modifications of DNA and proteins intimately associated with DNA, and in my case specifically, how the DNA is folded in the nucleus.
WHY IS THE FOLDING IMPORTANT?
If you stretched out the DNA from all of your body’s cells and lined it up end-to-end, it would cover the distance between earth and sun a hundred times. While this is certainly an intriguing tidbit of information, most importantly it goes to show how much information needs to
be compacted into a tiny nucleus.
HOW DOES IT ALL FIT?
The DNA is not randomly jammed into the nucleus. Instead, the packing follows a strict hierarchy, which is controlled by regulatory elements all over the genome. We want to understand how these regions control the packing of the genome and ultimately gene activity.
WHY WOULD THE PACKING INFLUENCE GENE EXPRESSION?
Control switches that regulate gene activity are found all over the genome, but they are not necessarily lined up next to the gene they control. By merely looking at the one-dimensional organization of the genome, we are unable to tell which gene a specific control switch is actually controlling. When a chromosome dynamically folds it brings distant control switches close to their targets, which are ready to respond to signals from control regions.
IS THE THREE-DIMENSIONAL ARCHITECTURE OF THE GENOME RELEVANT BEYOND ENSURING THAT IT ALL FITS?
It sounds like a very abstract concept when we talk about looking at the 3D structure of the genome, but that is not the case. We were able to show that activities of multiple genes that are implicated in disease are actually controlled by the three-dimensional folding. In our latest collaboration with Vijay (Associate Professor Pandurangan Vijayanand, M.D., Ph.D.) we found that the three-dimensional folding of a stretch of DNA is different in certain individuals who show increased susceptibility to asthma. Just five years ago, we wouldn’t have been able to measure such differences and we wouldn’t have considered such events to be relevant to gene regulation and disease.
HOW DO YOU ACTUALLY DETERMINE THE THREE-DIMENSIONAL STRUCTURE?
Traditionally, this was done through imaging, but imaging only gives you a limited number of reference points that tell you about the overall structure of a chromosome or the whole genome. Nowadays, you can take a snapshot of the nucleus by cross-linking cells, which chemically holds together the regions that are in close proximity. We then sequence the DNA to find out what the involved DNA regions are. We can now do this genome-wide by using many cells at once in order to look at all of the regions and how many times they get in touch with each other. However, these measurements don’t tell you the 3D structure or connections between genes and regulatory switches right away. That’s where the computational part comes into play and where I am having all the fun.
WHAT SET YOU ON YOUR PATH TO BIOINFORMATICS?
Biology was never a strong suit of mine. I was more interested in mathematics and abstract concepts such as computational complexity. Then I took a bioinformatics class from an inspiring young professor, and I had this ‘Aha!’ moment when I realized that many biological concepts could be modeled as computational problems—problems with a huge impact.
WHERE DID YOUR INTEREST IN THE ARCHITECTURE OF THE GENOME ORIGINATE?
Originally, I came from a computer science background, which by its very nature dealt with more abstract problems. When I started my postdoctoral training I wanted to be closer to data that had a direct relevance for understanding disease. I had several options, but the one project I was most interested in was the three-dimensional structure of the malaria parasite’s genome, which was then uncharacterized. That’s when I realized this is a lot of fun as well as an up-and-coming area of science and that there would be a lot of opportunities to develop new methods that will be widely used.
WHAT INSPIRES YOU AS A SCIENTIST?
As a computational scientist, I am wearing many different hats and I love them all: One day I might be working on malaria, the next day I might be working on cancer or asthma. Trying to catch up on the latest on all these different topics by immersing myself keeps things fresh and exciting. Plus, being able to come up with computational methods, which lots of people use in their day-to-day research, is very rewarding.
SCIENCE CAN BE ALL CONSUMING. HOW DO YOU KEEP A HEALTHY BALANCE?
[Laughs] I don’t know that I do. I love playing soccer and hiking and those really clear my mind. But with the new baby in the family it just doesn’t happen as much. Family
is very important for me and I make every effort to spend more time with them. Having a family has made me very flexible in terms of the places, positions, or ways that I can work in and be creative. I try to multifunction often and push the lines of what you can do with one hand when the other hand is holding a five-month-old.